Prepare, analyze, and visualize data with R
Arithmetic Artist
The R statistical programming language offers machine learning methods, dashboards, descriptive analyses, t-tests, cluster analyses, various regression methods, interactive graphs, and more. If you have ever wondered whether it would be worthwhile to immerse yourself in R, the following easy-to-grasp taster seeks to clarify who is likely to benefit.
Yesterday and Today
R [1] is based on the S programming language developed at Bell Laboratories in 1975-1976, which then split in the late 1980s and early 1990s into a commercial version named S-PLUS and the GNU project R. The name, which admittedly takes some getting used to, can be traced back to the first names of the developers, Ross Ihaka and Robert Gentleman, and alludes to the previous project name, as well.
R is often referred to as a programming environment, which is intended to emphasize the open package concept and to indicate that R differentiates itself from common monolithic statistical software. The basic functions are provided by eight packages included in the R source code. Additionally, many thousands of additional packages offer extensions.
In its early years, R was more of a niche player and was mainly used by statisticians and biometricians at universities. In the meantime, however, it has gained a firm place in the corporate world with the increasing entry of data science into many companies.
Syntax
An overview of the most important properties of its syntax facilitates any introduction to R. The R syntax is characterized by expressions and is case sensitive: An object named modelFit cannot be called as modelfit, for example. The assignment operator <- creates an object and points to the object that is assigned the content of an expression.
To assign the numbers from 1 to 5 to the vector numbers, use the following expression:
> numbers <- c(1, 2, 3, 4, 5)
The c() function – the c stands for "concatenate" – combines the individual elements listed in parentheses. An equals sign can be used as an alternative for assignments, in line with the standards of other programming languages. However, this practice is controversial in the R community. The assignment operator and equals sign also are not fully equivalent, because the latter can only be used at the top level.
Another member in the assignment operator group, <<-, is an extension of the assignment operator. Known as the superassignment operator, it can be used to assign values within functions in the global environment or to overwrite variables already defined there.
A special feature compared with common programming languages is indexing. To retrieve the first element of the numbers vector you would write numbers[1], which would return both the element at the first position of numbers and explicitly the index of that element ([1]).
As you will see from the code examples, no semicolon or the like is used to complete an expression. R uses line breaks for this purpose.
The interpreter anticipates you: If the end of a line obviously does not complete the expression (e.g., because a bracket is missing or the expression ends with a comma), the interpreter assumes that the expression continues in the next line and prompts you for the completion with a plus sign:
> rep(numbers, + each = 3) [1] 1 1 1 2 2 2 3 3 3 4 4 4 5 5 5
Comments in code are indicated by a hash,
# This is a comment
> names <- c("Anna", "Rudolf", "Edith",
+ "Jason", "Maria")
and can occupy a line of its own or be added to the end of a line.
Data Structures
The most important data structures in R are vectors, matrices, lists, and data frames. Vectors are unidimensional data structures and the smallest possible building block, because R has no scalars. For this reason, they are also known as atomic vectors. For example, an object containing only one string or one number is treated by R as a vector with a length of 1. R has five vector types:
logicalinteger-
double(numeric) charactercomplex
Vectors in R are characteristic, in that their elements all have to be of the same type. If you try to combine elements of different types (e.g., strings and numbers), you will not see an error message. Instead, R automatically converts all elements to the same class in a process known as coercion. The following example tries to create a vector from an integer, a logical constant, and a character. R automatically converts all elements to characters:
> misc <- c(43, TRUE, "Hello") > misc [1] "43" "TRUE" "Hello" > class(misc) [1] "character"
Data frames are table-like data structures in R and are used in almost every data analysis. Each column contains a vector; although all vectors have the same length, they can be of any type (Listing 1).
Listing 1: Data Frame
> data.frame(numbers, names)
numbers names
1 1 Anna
2 2 Rudolf
3 3 Edith
4 4 Jason
5 5 Maria
If you add an additional element to the names vector and again try to create a data frame from numbers and names, an error occurs because numbers has a length of 5 and names has a length of 6 (Listing 2).
Listing 2: Faulty Frame
> names[6] <- "Henry" > data.frame(numbers, names) Error in data.frame(numbers, names) : Arguments imply different number of lines: 5, 6
Strictly speaking, lists are also vectors, but they are recursive vectors. Any conceivable object can be components of a list, even lists themselves. The data structure list in R can be easily compared with a dictionary (dict) in Python or a structure (struct) in C. Lists in Python, on the other hand, are more similar to vectors in R, except R vectors can contain different data types.
Object Orientation
In R you can go quite a long way without ever knowing what a class is. Nevertheless, object-oriented programming is possible. R has three common systems of object orientation: S3 classes, S4 classes, and Reference Classes. The first two classes are the most common but implement a rather unusual concept. In contrast to common class systems, a generic function decides which method is called according to the class of the object to which it is applied (called method dispatch). Therefore, a function can be applied to different classes and return class-specific output.
For example, the summary() function applied to a dataset gives an overview of its dimension, the variables it contains, their classes, and so on. If you apply it to a numeric vector, R outputs minimum, maximum, and quantiles. S3 and S4 classes are fairly similar, but the S4 system is more formalized than the S3 system, and the method dispatch can take into account the classes of multiple input objects.
Reference Classes are more in line with the concept familiar from other languages that methods belong to classes rather than functions. The lack of significance of object-oriented programming in R can be explained by, among other things, the additional overhead it generally generates, rather than the benefits from the preparation and evaluation of data.
Standard Library and Extensions
R provides all functions through packages, some of which are already included in the R source code and are thus automatically installed and available. All other packages need to be downloaded and installed before their functionality is available. In addition to basic methods, the standard packages include functions for statistical data analysis, the creation of graphics, and sample datasets.
The packages base, datasets, and stats, among others, belong to the standard library. The base package provides – as the name suggests – really basic functions like mean(), length(), or print(). The datasets package contains small datasets that programmers can mess around with to test code and learn methods.
Also helpful is the stats package, which supports full-blown statistical analyses. Stats provides functions such as lm() (linear model), anova() (analysis of variance), and t.test() (significance difference of means test).
The main source for expanding packages is the Comprehensive R Archive Network (CRAN), a network of mirrored servers that provides developers with a platform on which to publish their packages. Packages available on CRAN must meet certain requirements primarily concerned with the architecture of the package, rather than the quality of the content. In addition to this official approach, packages can also be provided on GitHub or similar services.
To install the dplyr data manipulation package, for example, enter:
> install.packages("dplyr")
For the functions of the package to be usable, you first have to load the library:
> library("dplyr")
This step usually happens at the beginning of a session.
Graphics
The overhead for creating visually appealing graphics is greater in R than, for example, in Excel, but everything can be configured down to the last detail. The easiest way to generate graphics is to use the graphics package from the core distribution. The plot() function it includes automatically generates the appropriate graph type according to the input.
The strongest competitor to graphics is the slightly newer ggplot2 package. The syntax initially takes some getting used to; for example, the various elements like data, axes, and legends are connected by a plus sign. However, the learning curve is worth your while, because even with the default settings, ggplot2 graphics look much more professional than those from the graphics package.
A wide range of themes are easy to apply to the graphics, as well, making it possible to change subsequently the external appearance of a plot stored as an object. If the themes offered directly by ggplot2 are not to your taste, you can look around in the ggthemes package or even adjust the font, color, and size of the individual elements to suit your needs. Because it is not particularly difficult to define a theme yourself, you can create graphics that correspond exactly to the corporate design of an organization.
In the next example, the same plot is created first with graphics (Figure 1) and then with ggplot2 (Figure 2). To begin, you need a sample dataset with exactly one column (Listing 3) containing the responses by some imaginary people to a question about their favorite development environment in R (more about this later).
Listing 3: Sample Data Record
> dat <- data.frame(environment = c("RStudio", "Atom", "none", "RStudio", "Emacs", "RStudio", "RStudio",
+ "RStudio", "Emacs", "RStudio", "Atom", "RStudio", "RStudio"))
> plot(dat$environment)
> ggplot(dat, aes(x = environment)) + geom_bar()
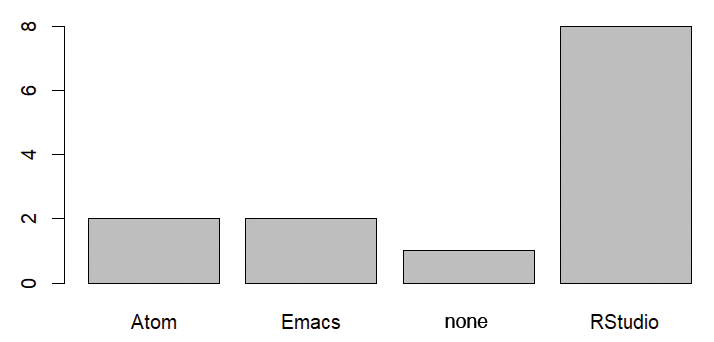
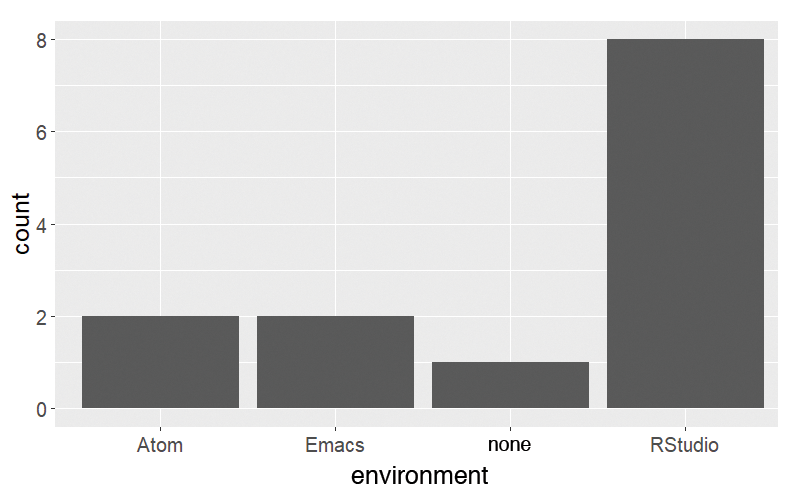
R base or Alternative Packages?
Typical applications in R can often be realized with the base package of the core distribution along with newer packages. In most cases, packages from the core distribution offer maximum stability, whereas some newer packages are much more convenient and elegant to use and have more sensible default values. However, some of these packages are still under development, so you should always keep in mind that a package update can change basic functionality. A quick look at the version number helps you avoid unpleasant surprises.
As already mentioned, you can create graphics with the graphics package from the core distribution or the newer ggplot2 (current version 3.1), which comes from the tidyverse collection of packages. Tidyverse is the home of a group of practical packages for data preparation, analysis, and visualization that follow a uniform interface.
Typical data preparation steps (changing data columns, calculating new columns, data aggregation, etc.) can be implemented both with the base package and with the tidyr (version 0.8) and dplyr (version 0.8) packages. The example in Listing 4 creates a small data frame with the columns a, b, and colour. R then computes the c and sum columns with the R base and dplyr packages.
Listing 4: Two Packages, One Task
> # base
> dat <- data.frame(a = c(10, 11, 12),
+ b = c(4, 5, 6),
+ colour = c("blue", "green", "yellow"),
+ stringsAsFactors = FALSE)
> dat$c <- 2 * dat$a
> dat$sum <- dat$a + dat$b
>
> # dplyr
> dat <- data_frame(a = c(10, 11, 12),
+ b = c(4, 5, 6),
+ colour = c("blue", "green", "yellow")) %>%
+ mutate(c = 2 * a,
+ sum = a + b)
The trademark feature of the dplyr package immediately stands out: The pipe operator (%>%) attaches expressions to each other like the links of a chain and forwards the expression to the expression on its right. Whereas complicated expressions often have to be nested in R base and then read from the inside out, these constructs can be broken down with dplyr and converted into logically consecutive steps.
The example in Listing 5 illustrates how the pipe operator improves the readability of the code. A logical vector, which also contains a missing value, is converted into a percentage. Even if the second example is made a bit longer by the pipe operator (and takes a few microseconds longer to execute), it is easier to see which arguments belong to which function.
Listing 5: Pipe Operator
> x <- c(TRUE, FALSE, TRUE, TRUE, FALSE, FALSE, TRUE, NA)
>
> # base
> paste0(round(mean(x, na.rm = TRUE), 2), "%")
[1] "0.57%"
>
> # dplyr pipe operator
> x %>% mean(na.rm = TRUE) %>% round(2) %>% paste0("%")
[1] "0.57%"
Development Environments
The R language has several development environments, of which RStudio (Figure 3) is by far the most common. In addition to the console, history, and editor with syntax highlighting, collapsing, and automatic indenting, the RStudio interface includes Git integration and helpful tools for the development of R packages.
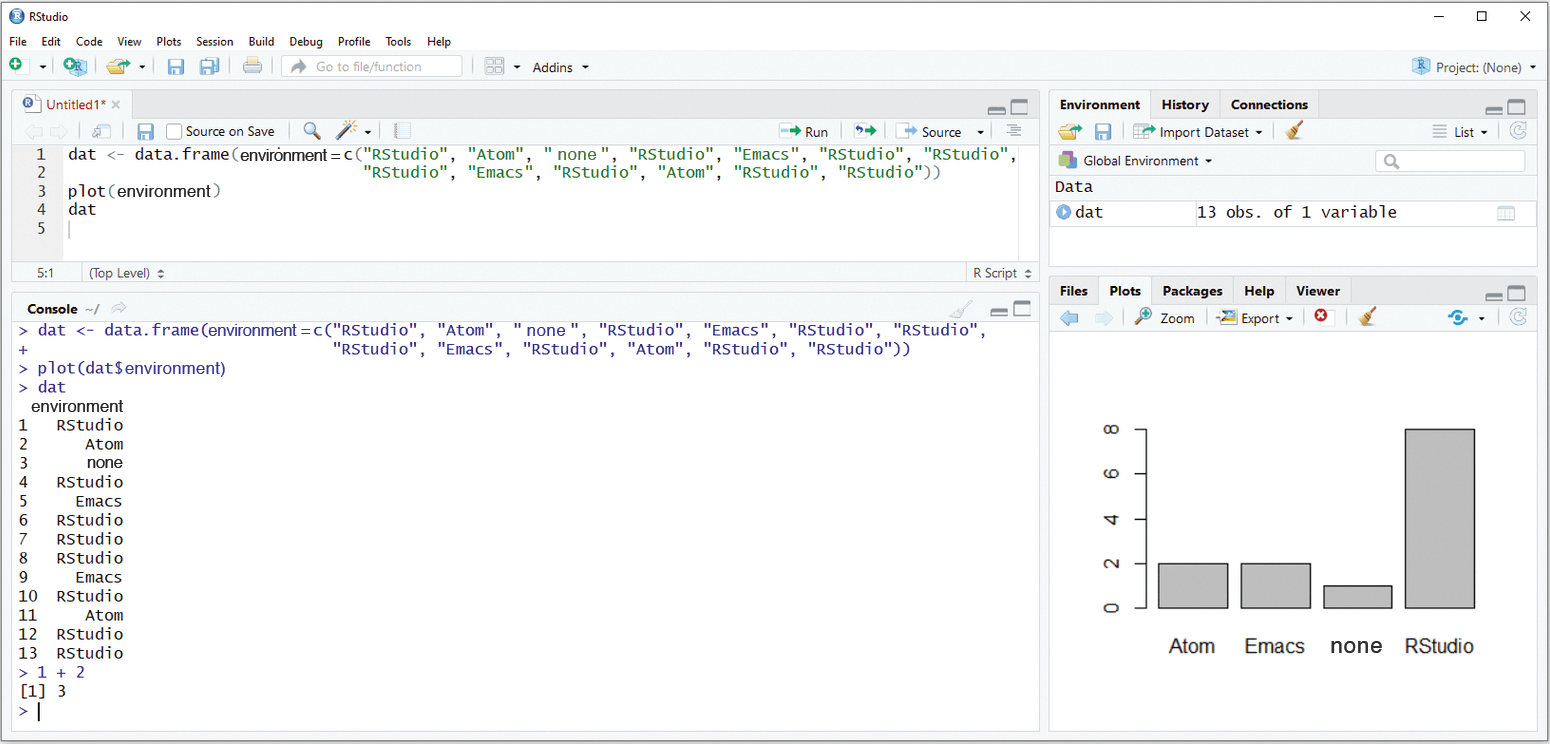
Alternatives to RStudio include Atom with the Hydrogen extension or Emacs with the ESS (Emacs Speaks Statistics) extension. Both open up more possibilities for individual configuration and expansion through plugins. They are suitable for different languages, so you have no need to change your editor. On the other hand, RStudio is often more convenient, especially for beginners, because it requires no configuration.
Strengths and Weaknesses
One of R's greatest strengths is certainly its overwhelming number of packages. You will find an R package suited to almost any application that has to do with data. However, the wide range of packages is also one of R's weaknesses, because it is often difficult to make a selection from among the large number available. Moreover, the stability and quality of rarely used packages sometimes does not meet the high level of the core distribution, not least because the authors often have only limited programming experience.
Further advantages and disadvantages are the result of R being an interpreted language. Because the code is not compiled first, code executes line by line in a terminal window; this method is the only way to enable interactive work and is almost indispensable. During explorative data analysis or the development of a statistical forecast model, you can look at your data, graphics, and results after every step. The intermediate step of error-free compilation would complicate data analysis enormously. The disadvantage, however, is that R is fairly slow in certain applications.
The problem of speed can be solved by parallelization, for which the parallel package is recommended. On Linux, the implementation is very uncomplicated, whereas the code for Windows is somewhat more complicated. Also, the memory requirement for parallelization on Windows is greater than for Linux, so the limiting factor is often not the number of cores, but memory (especially with large datasets).
Another way to accelerate code is to outsource performance-critical sections to C or C++; for example, the rcpp package is available for integrating C++ code. In practice, some users also follow the strategy of using R to develop a model interactively and finally implement the result for operational use in another language, such as Java.
Integration
R is as good as any other language in terms of database interfaces and the ability to integrate with other programming languages and programs. R communicates with MySQL, PostgreSQL, Oracle, and Teradata, among others, with native drivers, and with virtually any database (including Microsoft SQL Server) via ODBC (Open Database Connectivity) and JDBC (Java Database Connectivity).
Various solutions for Big Data applications can integrate R into Hadoop on your own hardware or on Amazon Web Services. R also has interfaces that link it with other programming languages: For example, R can exchange objects with Java (JRI (Java/R Interface) or rJava). The reticulate package lets you insert Python code into R, and you can reverse the process with the Python rpy2 package.
Support for R is growing for other statistical solutions and programs from the business intelligence sector, as well. For example, SPSS, SAS, MicroStrategy, and Power BI provide R interfaces and benefits from their wealth of functions.
For people who come from other programming languages (e.g., Python, C++, Java), R takes some getting used to. As mentioned in an earlier example, you do not have to specify type explicitly when creating a new object. R automatically converts data to another type under certain conditions. For example, when two objects of different types are compared, they are converted to the more general type (e.g., 2 == "2" leads to the result TRUE), so for full control, it makes more sense to use the identical() function for comparisons. If you are not aware of this feature, you could see unexpected results.
Loops slow down code, so instead, R developers should choose a vectorized approach. For example, the apply() functions let you apply the same function to all elements of a vector, matrix, list, and the like. For programming enthusiasts with extensive experience in other languages, this can mean some acclimatization.
Last but not least, like many older languages, R has some peculiarities that can only be explained historically. For example, when importing datasets, string variables are stored by default as factor (categorical data with a limited number of values) and not as simple characters (string data). At the time R was created, analysis of categorical variables was more common than analysis of real text, so strings at that time usually reflected categories. Today, therefore, the argument
stringsAsFactors = FALSE
usually has to be passed in when reading strings (e.g., with read.csv()).
No uniform naming scheme has been able to assert itself thus far, either: Function names of the statistical package from the core distribution contain dots (t.test()), underscores (get_all_vars()), lowercase (addmargins()), and CamelCase (setNames()).
From Science to Business
R clearly has its roots in the scientific community; it was developed at a university and is still widely used in science. Scientific publications that include new statistical methods often include an R package that implements the method.
In addition to R, of course, scientists also like to use other tools when they prepare and analyze data. Some of these tools offer graphical user interfaces but limited functionality (e.g., SPSS [2], which is very popular in psychology and social sciences). Like SPSS, other tools are subject to a fee, including Stata [3] (econometrics) and SAS [4] (biometrics, clinical research, banking).
Python is widespread in the computer science environment and especially in machine learning, where it leaves hardly anything to be desired, even compared with R. In terms of statistical applications, the main difference between the two languages is the variety of packages available in R. In Python, data science applications are concentrated in a smaller number of more consolidated packages, but more specialized or newer methods are often not available.
Whether you prefer R or Python depends in the end on the technical background of the team. If you have a background in computer science, you might already have knowledge of Python and want to use it in a professional context. People with a mathematical or scientific background, on the other hand, are more likely to have experience with R.
R owes its leap from scientific software into the business world to the proliferation of data. Companies seek to profit from their data and optimize their business processes. The employees of a company faced with the task of analyzing data, often use Microsoft Excel. As a very powerful program, Excel can also meet many requirements. However, if more complex data processing becomes necessary or statistical models need to be developed, users can easily reach the limits of Excel.
Getting started with R offers more advantages than just the multitude of methods for data preparation and analysis. For example, R Markdown can be used to create reproducible and parameterized reports in PDF, Word, or HTML format. The use of LaTeX elements in R Markdown reports opens countless possibilities, such as the adaptation of reports to a corporate identity [5]; however, no LaTeX knowledge is required for working with R Markdown.
The Shiny package makes it possible to develop interactive web applications [6], which means R can cover tasks that would otherwise require tools such as Tableau [7] or Power BI [8]. However, automating processes with the help of R scripts already offers enormous benefits for many newcomers compared with the repeated execution of the same click sequence.
Regression Analysis Example
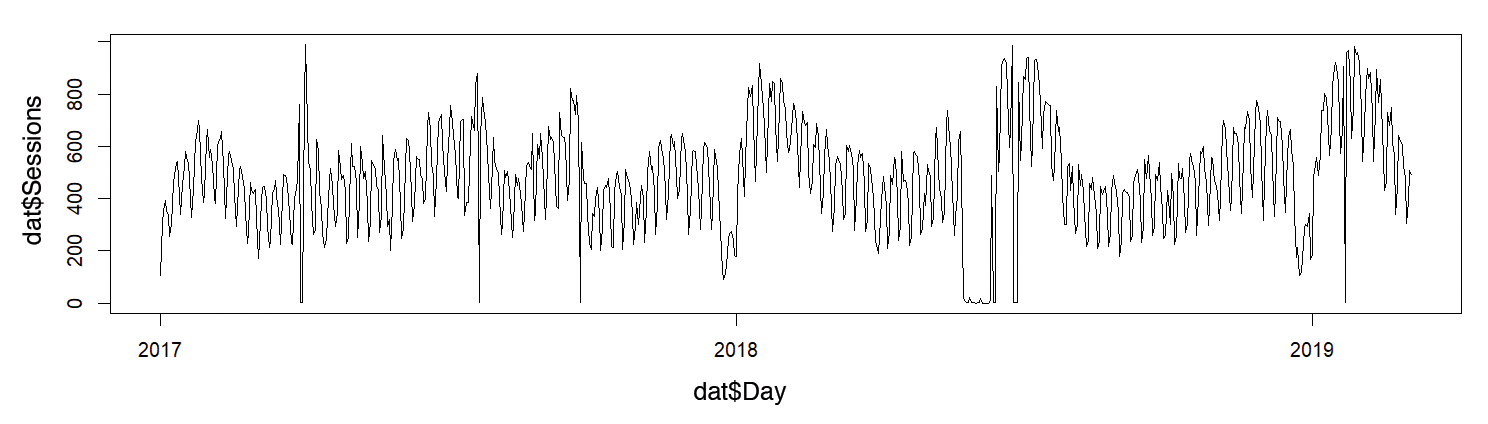
The final example reads Google Analytics data from a CSV file that has the number of sessions on the INWT Statistics [9] website since the beginning of 2015 (Figure 4). First, the script filters and prepares the data (Listing 6); then, with the data for 2017 and 2018, the code models the number of sessions, depending on the month and day of the week, and initially withholds the first few weeks of 2019 as test data.
Listing 6: Time Series Analysis
> # Load required packages
> library(dplyr)
> library(ggplot2)
> library(ggthemes)
>
> # Read data from csv file
> dat <- read.csv("data/Google_Analytics_Sessions.csv", stringsAsFactors = FALSE)
>
> # First view of the dataset
> str(dat)
'data.frame': 794 obs. of 2 variables:
$ Day : chr "01.01.17" "02.01.17" "03.01.17" "04.01.17" ...
$ Sessions: num 105 304 355 394 356 338 255 302 461 489 ...
>
> # Convert the date stored as character to a date (with base package)
> dat$Day <- as.Date(dat$Day, format = "%d.%m.%y")
>
> # Graphical representation of the time series with the built-in graphics package (Figure 5)
> plot(dat$Day, dat$Sessions, type = "l")
>
> # Data preparation with dplyr
> datPrep <- dat %>%
+ mutate(
+ # Set values below 50 to NA, because this is known to be a measurement error.
+ Sessuibs = ifelse(Sessions < 50, NA, Sessions),
+ # Determine weekday
+ wday = weekdays(Day),
+ # Determine month
+ month = months(Day)
+ )
>
> # Split into training and test data
> datPrep$isTrainData <- datPrep$Day < "2019-01-01"
>
> # Simple linear model. The character variables "wday" and "month" will be
> # automatically interpreted as categorial variables.
> # The model only uses the data up to the end of 2018, i.e. for which the variable
> # isTrainData has the value TRUE
> mod1 <- lm(data = datPrep[datPrep$isTrainData, ], formula = Meetings ~ wday + month)
>
> # Model summary
> summary(mod1)
Call:
lm(formula = Sessions ~ wday + month, data = datPrep[datPrep$isTrainData, ])
Residuals:
Min 1Q Median 3Q Max
-464.80 -61.88 -6.52 62.38 479.19
Coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept) 517.340 18.270 28.316 < 2e-16 ***
wdayThursday -31.046 16.119 -1.926 0.0545 .
wdayFriday -116.981 16.122 -7.256 1.08e-12 ***
wdayWednesday -2.817 16.111 -0.175 0.8612
wdayMonday -7.527 16.030 -0.470 0.6388
wdaySaturday -262.736 16.164 -16.255 < 2e-16 ***
wdaySunday -202.964 16.077 -12.624 < 2e-16 ***
monthAugust -16.028 20.777 -0.771 0.4407
monthDecember -6.106 20.768 -0.294 0.7688
monthFebruary 109.349 21.304 5.133 3.72e-07 ***
monthJanuary 133.990 20.770 6.451 2.10e-10 ***
monthJuly 211.030 20.849 10.122 < 2e-16 ***
monthJune 167.145 22.755 7.345 5.85e-13 ***
monthMay 24.528 21.411 1.146 0.2524
monthMarch -10.467 20.858 -0.502 0.6159
monthNovember 99.135 20.943 4.734 2.68e-06 ***
monthOctober -27.591 20.770 -1.328 0.1845
monthSeptember 53.475 21.030 2.543 0.0112 *
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
Residual standard error: 114.2 on 685 degrees of freedom
(27 observations deleted due to missingness)
Multiple R-squared: 0.5528, Adjusted R-squared: 0.5417
F-statistic: 49.81 on 17 and 685 DF, p-value: < 2.2e-16
> # Calculation of the values predicted by the model (for the whole period)
> datPrep$pred <- predict(mod1, newdata = datPrep)
>
> # Graphical representation of true and predicted values with ggplot2
> p <- ggplot(datPrep) +
+ geom_line(aes(x = Day, y = Sessions, linetype = !isTrainData)) +
+ geom_line(aes(x = Day, y = pred, colour = !isTrainData)) +
+ scale_color_manual(values = c("#2B4894", "#cd5364"), limits = c(FALSE, TRUE)) +
+ labs(colour = "Test period", linetype = "Test period")
> p # Equivalent to print(p)
>
> # Alternative model with splines
> library(mgcv)
> library(lubridate)
> datPrep$dayOfYear <- yday(datPrep$Day)
> datPrep$linTrend <- 1:nrow(datPrep)
> mod2 <- gam(data = datPrep[datPrep$isTrainData, ],
+ formula = Sessions ~
+ wday + linTrend + s(dayOfYear, bs = "cc", k = 20, by = as.factor(wday)))
> summary(mod2)
Family: gaussian
Link function: identity
Formula:
Sessions ~ wday + linTrend + s(dayOfYear, bs = "cc", k = 20,
by = as.factor(wday))
Parametric coefficients:
Estimate Std. Error t value Pr(>|t|)
(Intercept) 532.0423 12.3948 42.925 < 2e-16 ***
wdayThursday -32.2007 14.1368 -2.278 0.0231 *
wdayFriday -119.6377 14.1354 -8.464 < 2e-16 ***
wdayWednesday -4.9203 14.1359 -0.348 0.7279
wdayMonday -5.8354 14.0639 -0.415 0.6784
wdaySaturday -265.6973 14.1727 -18.747 < 2e-16 ***
wdaySunday -204.1019 14.1028 -14.472 < 2e-16 ***
linTrend 0.1254 0.0205 6.118 1.71e-09 ***
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
Approximate significance of smooth terms:
edf Ref.df F p-value
s(dayOfYear):as.factor(wday)Tuesday 15.32 18 6.413 < 2e-16 ***
s(dayOfYear):as.factor(wday)Thursday 12.97 18 3.902 1.89e-10 ***
s(dayOfYear):as.factor(wday)Friday 12.15 18 3.953 4.59e-11 ***
s(dayOfYear):as.factor(wday)Wednesday 13.85 18 5.314 9.31e-15 ***
s(dayOfYear):as.factor(wday)Monday 16.64 18 8.382 < 2e-16 ***
s(dayOfYear):as.factor(wday)Saturday 11.29 18 3.307 3.00e-09 ***
s(dayOfYear):as.factor(wday)Sunday 12.92 18 4.843 1.02e-13 ***
---
Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
R-sq.(adj) = 0.648 Deviance explained = 69.9%
GCV = 11749 Scale est. = 10025 n = 703
> datPrep$pred2 <- predict(mod2, newdata = datPrep)
> ggplot(datPrep) +
+ geom_line(aes(x = Day, y = Sessions, linetype = !isTrainData)) +
+ geom_line(aes(x = Day, y = pred2, colour = !isTrainData)) +
+ scale_color_manual(values = c("#2B4894", "#cd5364"), limits = c(FALSE, TRUE)) +
+ labs(colour = "Test period", linetype = "Test period")
In the Estimate column of the model summary, you can see that Saturdays and Sundays have very few website visits and that June and July are particularly strong months. Interpretation is always by comparison with the Tuesday or April reference class, which is set automatically. All told, the model can already explain greater than 50 percent of the variation (Multiple R-Squared = 0.5528).
The example then computes the values predicted by the model, including the first weeks of 2019, so you can investigate how well the model works with new data that was not used for the estimate. Finally, the actual number of sessions and the number predicted by the model are displayed graphically (Figure 5).
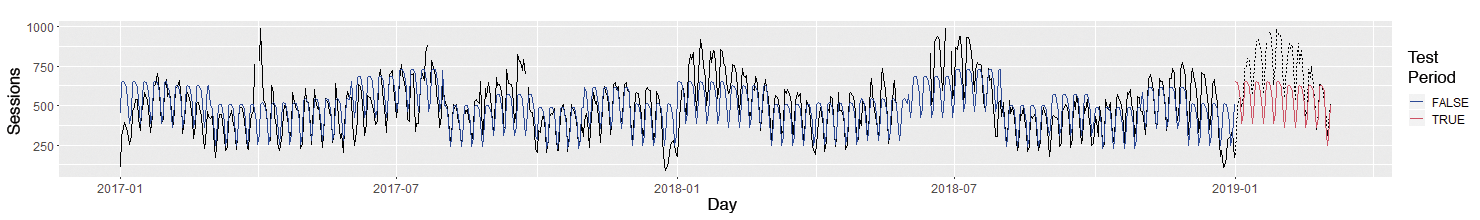
The figure clearly shows that the very simple model still has some weaknesses; for example, it does not predict the slump around Christmas and New Years. Another step checks whether adding holiday variables improves the model – visually or by using model measures such as R-squared (ideally on test data that was not used to estimate the model).
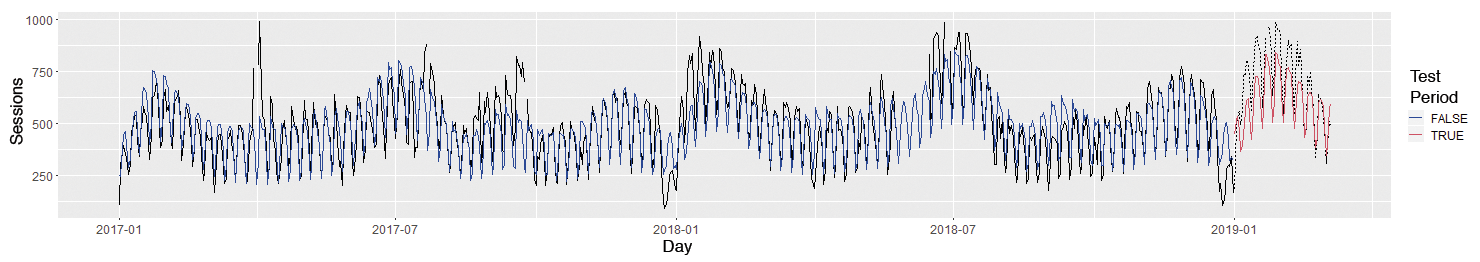
The model improves significantly if you map the annual trend nonlinearly with a spline rather than the month variable and add a linear trend (Figure 6). Whether the model will deteriorate if the days are combined into "weekday vs. weekend/holiday" instead of considering them individually is open to question.
Conclusions
In particular for universities, but also for enterprises, the often crucial point is that R can be used free of charge, in contrast to commercial solutions for data analysis. It also supports all common operating systems (Linux, Windows, macOS). The range of free help resources, including manuals, tutorials, and blogs [10], is very extensive. Community support is also very good, along with additional fee-based support from third-party providers and numerous training offerings. The large developer community ensures that the project is actively developed in the medium and long term.
For beginners without any programming experience, the barriers to getting started with R are fairly low; in fact, you can get started quite quickly and learn the basics required for statistical analysis within a few days. Reading a dataset and computing a regression model takes just two lines of code, and you do not need to deal with concepts such as object-oriented programming.
To sum things up, R is suitable for any application scenario in which statistics software is used regularly. The main arguments – besides the elimination of licensing costs for the software itself – are, above all, the breadth of supported methods, the enormous flexibility, and good automation. The active community offers support and guarantees the long-term development of the software.
Although data analysis can in principle be performed with any programming language, R was developed specifically for this purpose, and many common methods are already implemented. Only if very large datasets have to be analyzed in a limited time on comparatively weak hardware, if users use statistics software only sporadically, or if you only need very special methods is the use of R questionable.
If you want to keep track of the latest developments or want to chat with a member of the Core Team, you can visit an R conference like useR! [11] or the European R Users Meeting (eRum) [12]. Many larger cities have regular R meetups [13].
